Searching
To search for an ortholog group you can type a gene identifier or a gene description in the search box, that is located at the navigation bar, and press either the search button or the enter key. After that, you will get redirected to the tree view page.
Browsing
It is also possible to retrieve trees by using the browsing feature. On the page the species tree used by the Hieranoid algorithm is displayed. Here you can select species by typing their names in the search box provided on the page. With clicking the search button, the last common ancestor for the given values and its children nodes are highlighted.
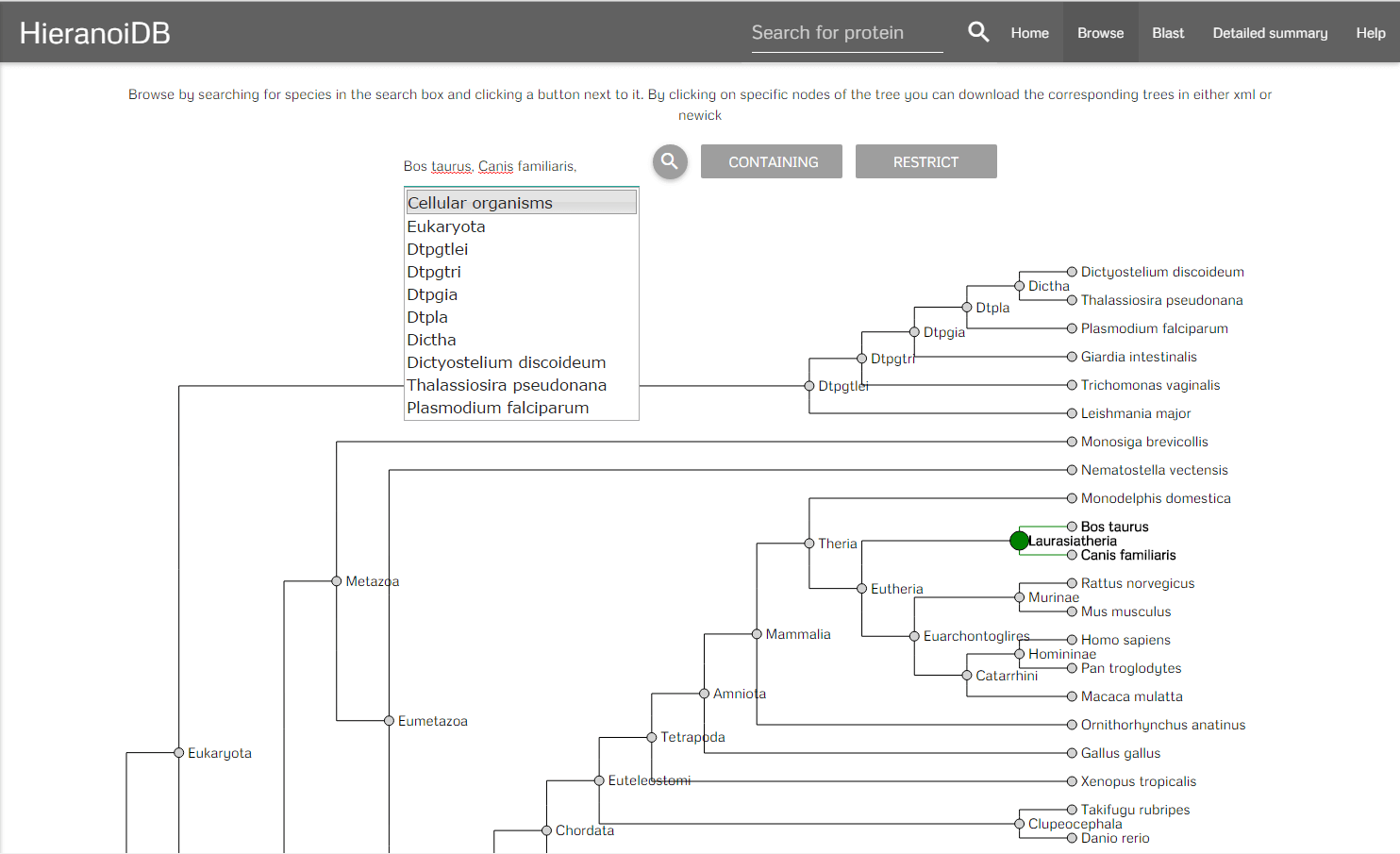
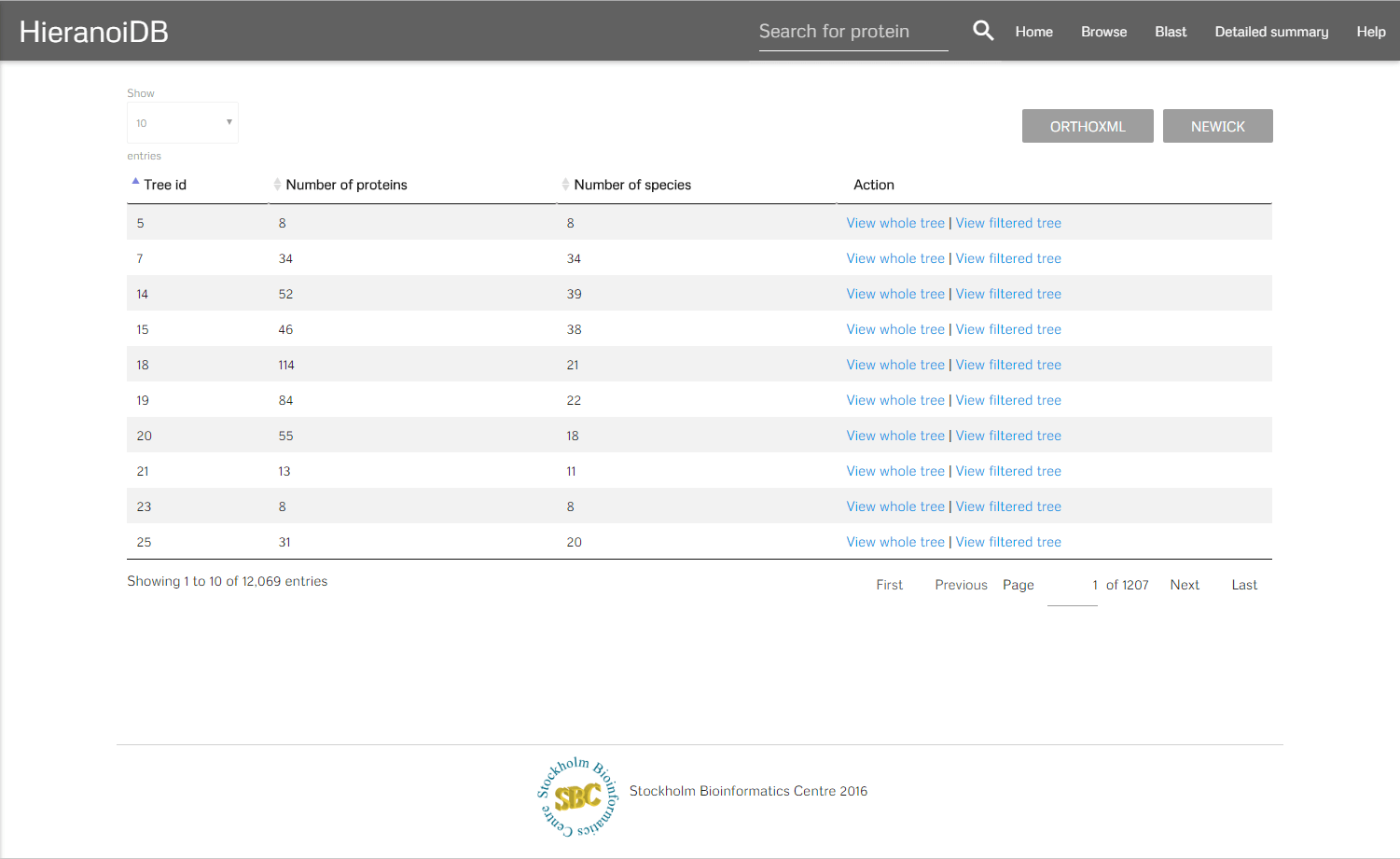
After selecting a set of species there are two buttons that can be clicked. On clicking the "containing" button you will get redirected to a list of trees which contains all trees that include any of the selected species. On clicking the "restrict" button you will get a list of trees which contains all trees that are exclusive to the set of species which was selected. It is also possible to click on a node and get directly to the corresponding trees. In the resulting list it is possible to display the individual trees by clicking on a link, you can either show the full tree or a tree that is limited to the previously selected species.
Blast
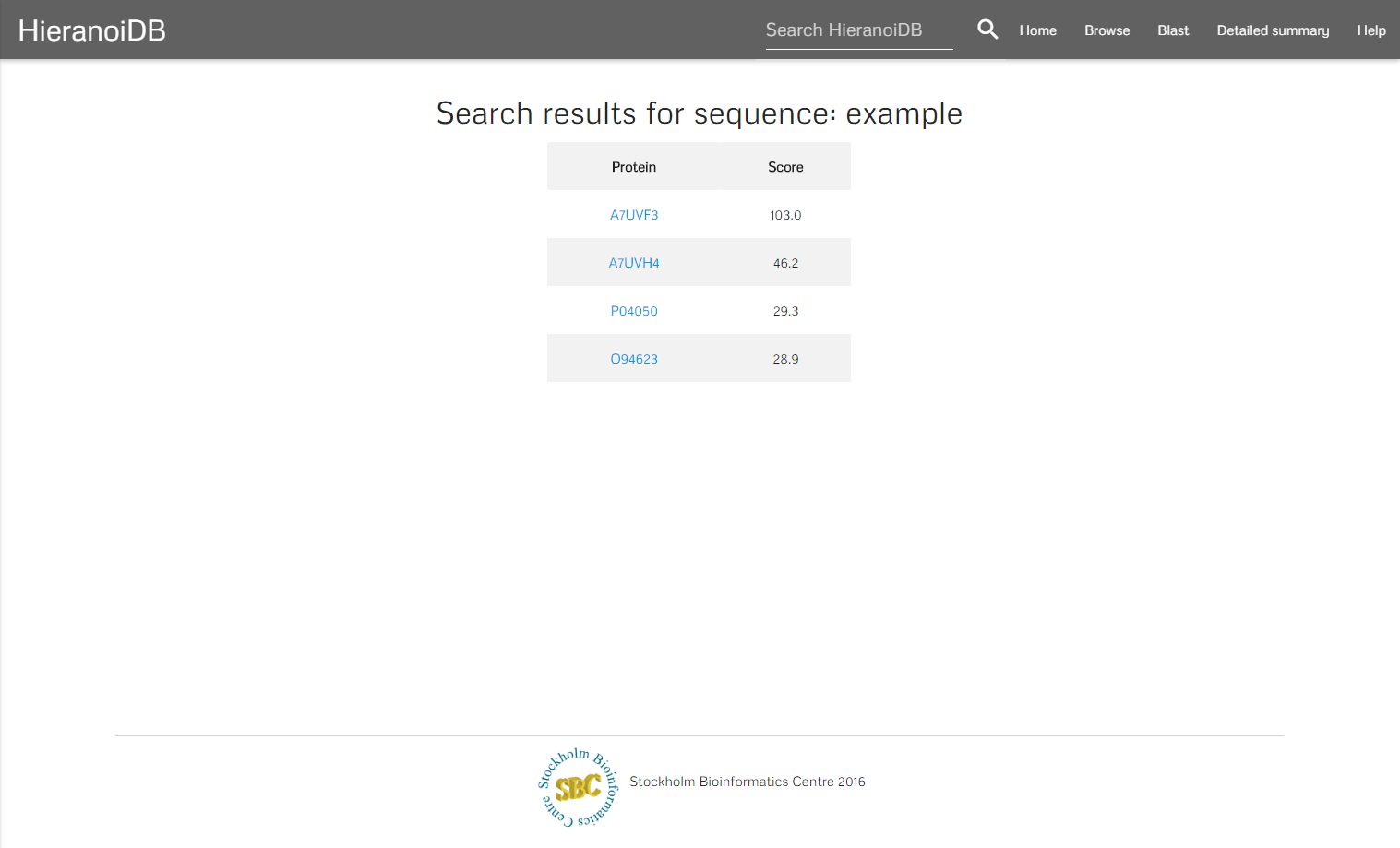
A further search possibility is to query the database using Protein-Protein BLAST 2.3.0+, which retrieves a list of proteins with similar sequences for any number of entered protein sequences in FASTA format.
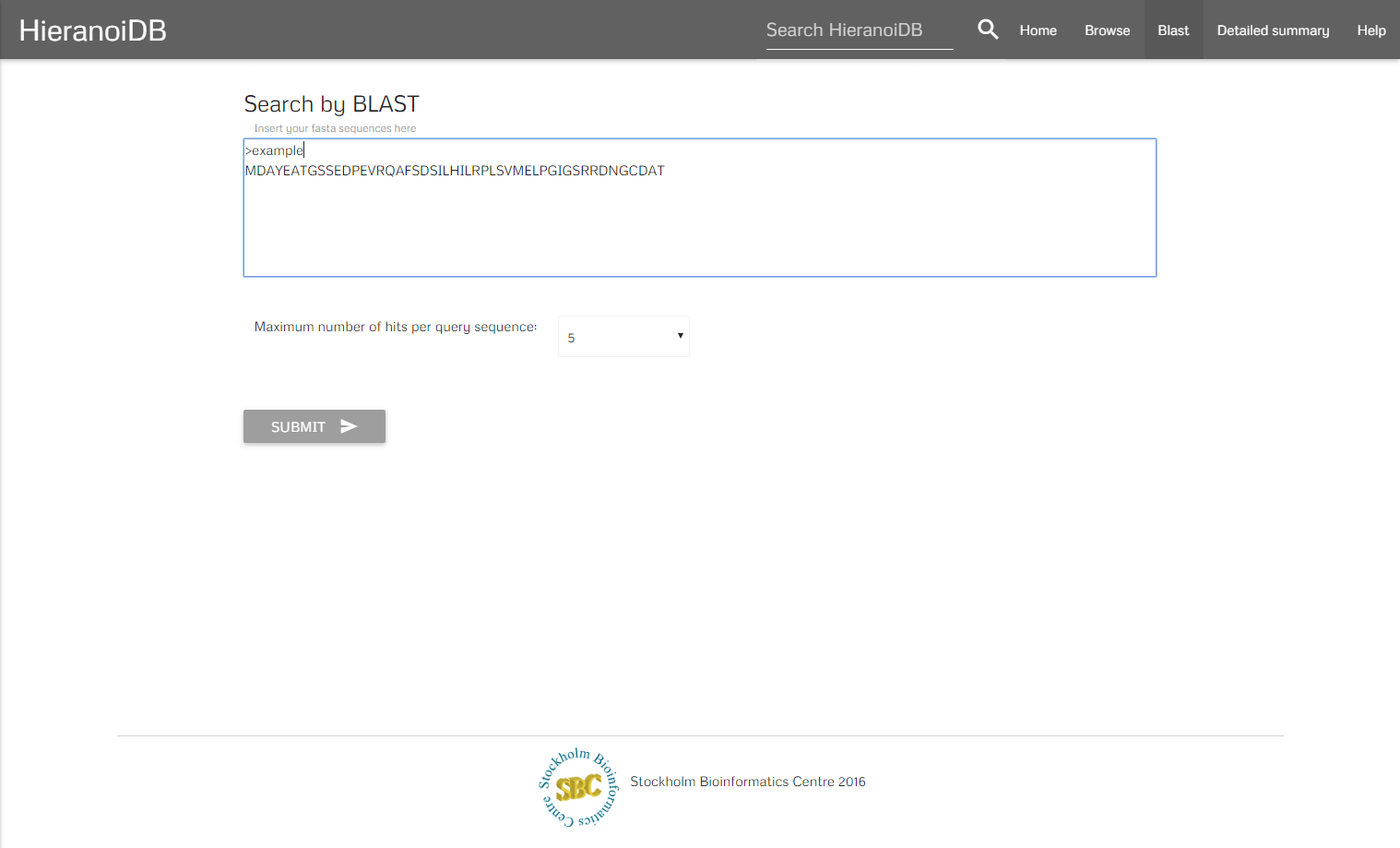
The algorithm retrieves a list of genes that are similar to the entered sequences. By clicking on a gene identifier you will get redirected to the corresponding tree.
Tree view
After a tree was found by using one of the above methods it is displayed and you can interact with it.
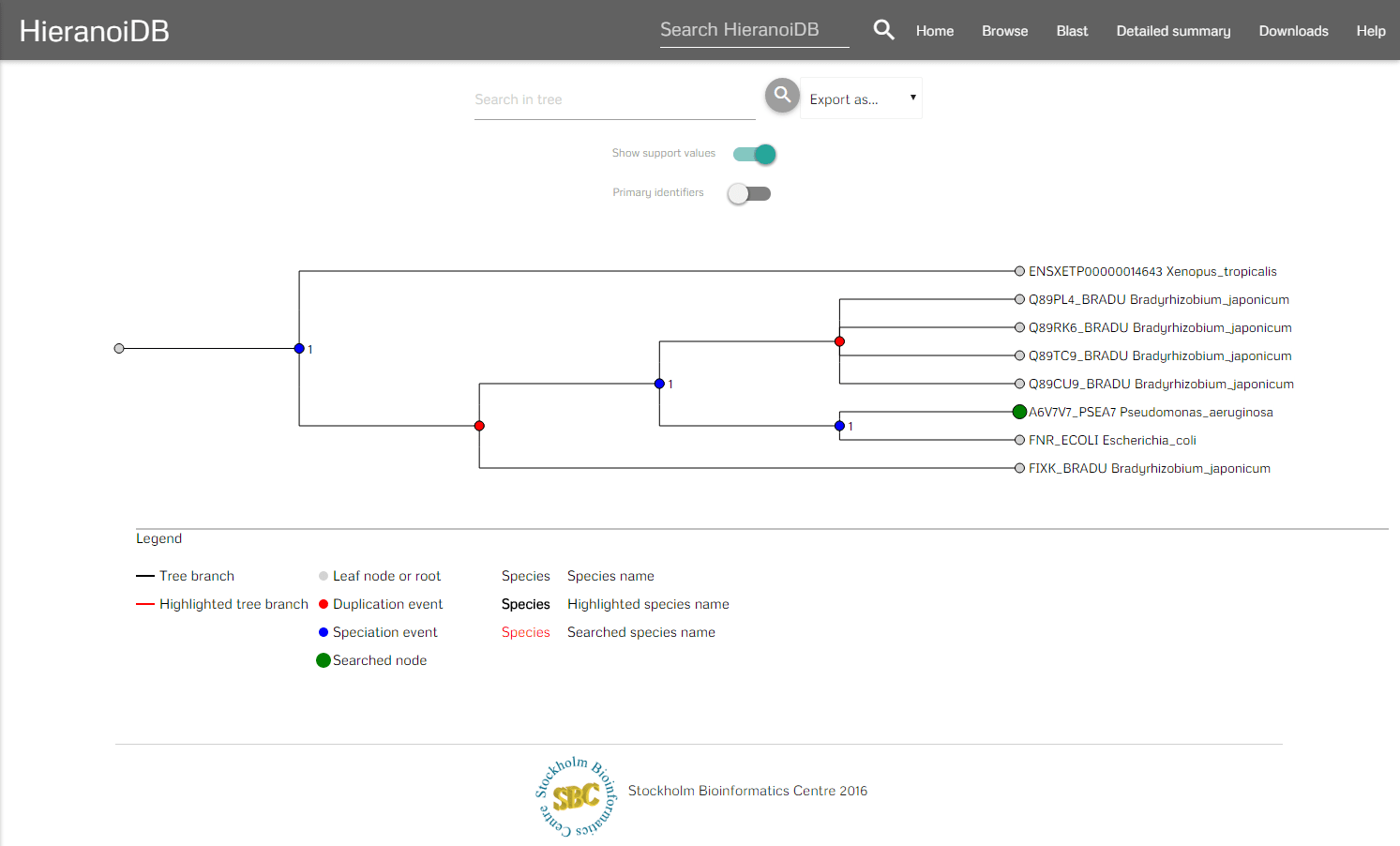
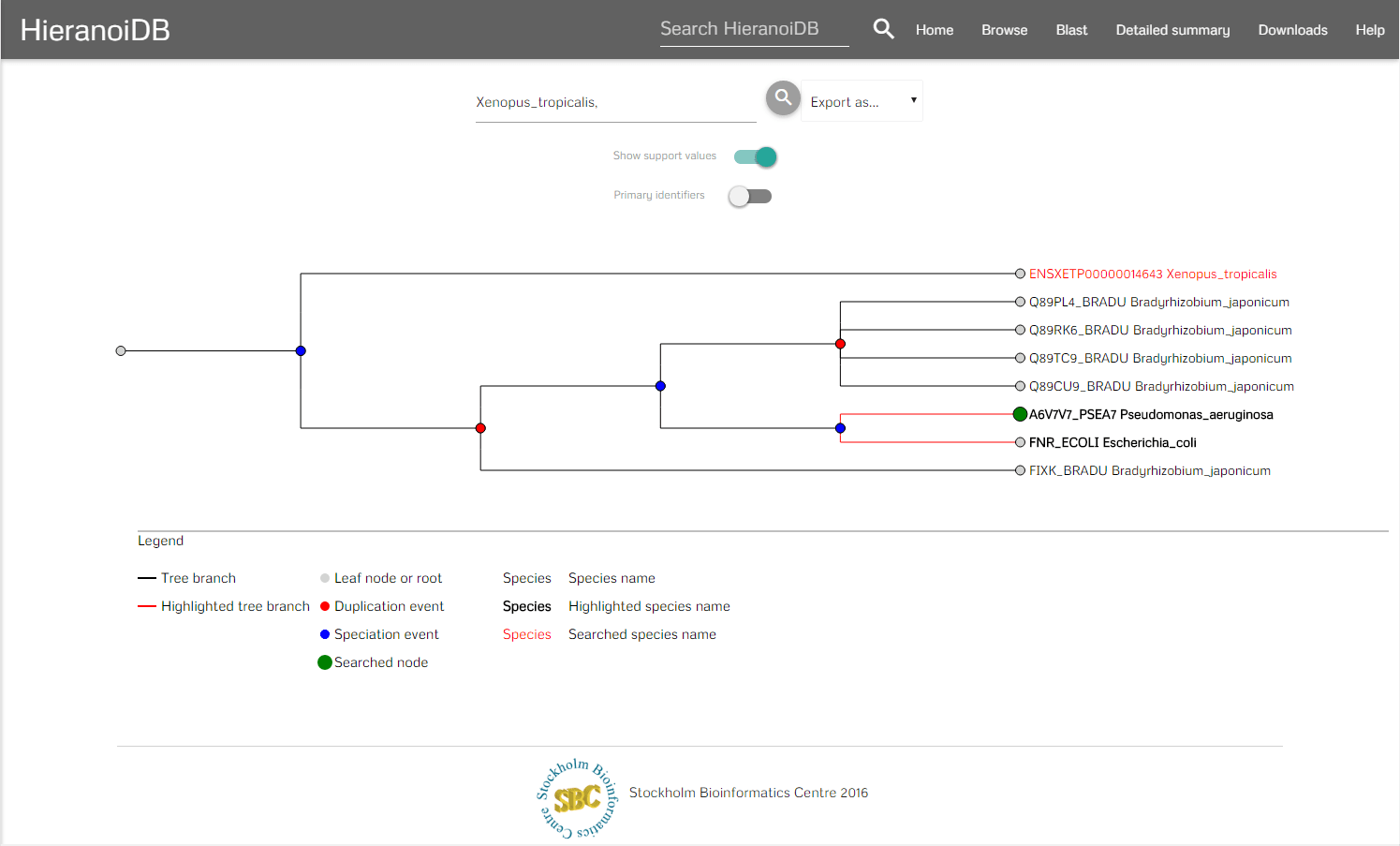
In the menubar you can search for certain nodes (which marks the corresponding labels as red) and download the tree in different formats e.g., orthoxml, newick. Furthermore, it is possible to download the tree as Scalable Vector Graphics (SVG) and the tree's gene sequences can be exported as a fasta file aswell. It is also possible to hide the support values in the tree and to show the original identifiers by using the provided switches. To interact with the tree you can click on nodes and retrieve their information as a tooltip. In the tooltip you are provided some information of the node (species, description, etc.) and you can further interact with the tree - here you can collapse branch nodes and highlight subtrees (which colors the branches red and changes the font size of the labels). If the tree has been changed, then also the SVG download will be adapted accordingly to contain the modified tree.
Citation
Kaduk M, Riegler C, Lemp O, Sonnhammer EL. HieranoiDB: a database of orthologs inferred by Hieranoid. Nucleic Acids Res. 2017, 45(Database issue), D687-D690. PMID: 27742821 [Link]
Contact
Do not hesitate to contact us for feedback, questions and suggestions.
E-Mail: mateusz.kaduk@scilifelab.se
Erik Sonnhammer
Mateusz Kaduk
Christian Riegler
Oliver Lemp
The Sonnhammer Lab at the Stockholm Bioinformatics Centre